1 - RNA Transcription Flashcards
(29 cards)
Major Diff Between RNA and DNA
DNA RNA
Function is genetic info genetic expression
Sugar is 2-deoxyribose ribose
Pyrimidine bases are cytosine cytosine and uracil
and thymine
Usually double stranded single stranded
Size can exceed 1 mil base pairs 60-20,000 bp
No enzymatic function ribozyme
Conformation is restrained by Greater flexibility
being double stranded
- Ribozyme = RNA that is an enzyme ; can also catalyze its own replication also found in our own body
- Example of single stranded RNA: in case of viral infection, signals for interferons.

Major Diff Between RNA and DNA
DNA RNA
Found in nucleus nucleus and cytoplasm
Deoxyribose sugar is not Ribose sugar is more
very reactive. Stable in alkaline reactive. Not stable in
conditions. Small grooves alkaline conditions. Has
where nucleases attach. larger grooves.
Chromosomal DNA is Less sensitive to UV rays
primarily B form. Sensitive
to UV rays.

Types of RNA in Eukaryotes
- Messenger RNA conveys genetic information from the DNA to the protein. Makes up about 5% (subsequently small percentage) of the total RNA in the cell
- Ribosomal RNA …18S constitutes part of the small subunit of the ribosome
- 28S,5.8S, 5s constitute part of the large subunit of the ribosome ( we do not need to know the numbers or percentage)
- Make up approximately 80% of the total RNA in the cell
- Transfer RNA …Serves as adaptor molecule in protein translation
- Makes up about 15% of the total RNA in a cell
- There are 32 different types of tRNA in most eukaryotic cells
- Contain no more than 93 nucleotides (very small)
-
Small nucleolar RNA are found in the nucleolus, where they are involved in:
• Synthesis of ribosomes
• Chemical modifications of other RNA molecules
• Alternative splicing
• Synthesis of telomeres - MicroRNA: contains about 20 nucleotides, are involved in regulating the levels of expression of certain genes
-
XIST RNA: large molecules of RNA responsible for inactivation of X chromosome in females
- Two X’s exist early in development but one of them is inactivated forever; considered to be a backup system where having two X’s increase the likelihood of at least one being a viable/”healthy” gene.
- Small nuclear RNA….Are involved in processing of other RNA’s (e.g., are found in splicesomes)
Transcription
Prokaryotic and eukaryotic RNA’s
- Just know they have different sizes, shapes, and functions
- Ribosomes also consist of different proteins on top of rRNA’s

Ribosomes
- Cells can have many ribosomes, which is the reason why rRNA can account for 80% of the total RNA in a cell (It’s been reported that a very active mammalian cell can synthesize almost over 1000 rRNA molecules each second, each of which associates with 150 different snoRNA’s , which subsequently combine with 80 different proteins to create the ribosomes; especially greater for more active cells).
- The genes for 28S, 18S, and 5.8S rRNA are usually
repeated in tandem clusters; they are transcribed by RNA Polymerase I as a single 45S rRNA precursor, which is subsequently processed (chopped off) to produce the 3 rRNA mature transcripts; only rRNA’s go through this NOT mRNA’s
Eukaryotic rRNA and its
posttranscriptional processing

tRNA
- has double stranded character
- tRNA has a cloverleaf structure (in 3D sense) with a significant amount of tertiary structure composed of stems and loops
- Many of the nucleotides are modified (Modified nucleotides can include ribothymidine, pseudouridine, dihydrouridine, etc.)
- Eukaryotic tRNA genes are transcribed by RNA
Polymerase III, and then undergo additional processing, which may includes the following: cleavage of the additional 5’ sequence, intron splicing, addition of the CCA sequence at the 3’end, and base modification.- No need to know the RNA pol # except for Pol II, which is associated with mRNA.
- Both mRNA and tRNA undergo significant post translational processing.

Post-transcriptional
Modification of RNA
- Ribosomal and Transfer RNA both go through extensive post-transcriptional processing
- Pos-transcriptional processing of tRNA:
- A 16 nucleotide sequence at the 5’ end is cleaved by RNAse P in pre-tRNA
- A 14-nucleotide intron in the anticodon loop is removed by splicing in pre-tRNA
- Many bases are converted to characteristic modififed bases
- Uracils at the 3’ end are replaced by the CCA sequence found in mature tRNAs

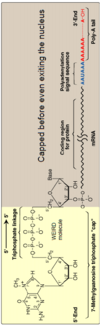
Eukaryotic mRNA
- Only very specific portion of Eukaryotic mRNA actually codes for protein i.e. the Coding Region
- Eukaryotic mRNA contains:
-
A cap (a methylated guanosine triphosphate attached through a 5’ to 5’ linkage to a the hydroxyl on the ribose at the 5’ end of the mRNA)
- It doesn’t look like anything else and very unique thus cannot be processed by nucleases; protection from degradation
- NOT something coded by DNA
-
A cap (a methylated guanosine triphosphate attached through a 5’ to 5’ linkage to a the hydroxyl on the ribose at the 5’ end of the mRNA)
-
Poly (A) tail ( up to 200 adenine molecules at the 3’ terminus of the mRNA)
- Simply added on; NOT something that originated from DNA
- Both the cap and the poly A tail serve to stabilize the mRNA against exonucleolytic degradation and participate in polypeptide chain initiation; play a role in translation and stabilization
- The cap is added immediately after synthesis, followed by the addition of the poly A tail.
- Neither the cap or tail are coded for in the DNA, protects it so nucleases won’t attack it

Base pairing between DNA and RNA
- Antiparallel, complementary base paring between DNA and RNA

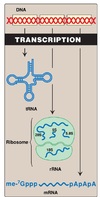
RNA Synthesis
- RNA polymerase can initiate the synthesis of the RNA chain without the need for a primer.
- The essential precursors for the synthesis of the chain are CTP, UTP, GTP, and ATP
- The DNA template is copied in the 3’ to 5’ direction
- The RNA chain is synthesized in the 5’ to 3’ direction
- Local unwinding of the DNA occurs after binding of the RNA polymerase; the template strand (vs. Non-template) is subsequently transcribed; RNA will ook very much alike non-template strand of DNA
Prokaryotic RNA polymerase
- The prokaryotic RNA polymerase consists of the core enzyme plus a dissociable sigma factor. The sigma factor recognizes transcriptional start sites, and the core enzyme polymerizes nucleotides into RNA. (Except for the short RNA primers associated with DNA synthesis), there is only one species of prokaryotic RNA Polymerase.
- Omega imparts specificity on what to transcribe and where to the polymerase should bind and work.

Prokaryotic promoter region
- Polymerase will recognize and bound upstream of +1 in the DNA strand.
- There is no “zero”th bp; transcription starts at +1
- Anything upstream of +1 is given Negative numbers
- No need to memorize the specific bp sequence but know their names and whereabouts
- Pribnow box from -7 to -12 (around -10)
- -35 sequence from -32 to -37 (around -35 hence the name)
- Sigma recognizes the -35 sequence and Pribnow box.
- If the consensus sequence are close enough, they are fine, doesn’t have to be 100% perfect
- However, there is a possibility of going much slower and lesser quality translation.
- The prokaryotic promoter serves as the binding site for the RNA polymerase.
- The prokaryotic promoter consists minimally of the -35 site ( consensus sequence is TTGACA) and the -10 site ( Pribnow box).
- The nucleotides in the Pribnow box dissociate (Pribnow box consensus sequence is TATAAT), allowing the RNA Polymerase to separate the DNA double helix and to initiate transcription.
- The less the specific recognition sequences resemble the consensus sequence, the slower the transcription.

Local unwinding of DNA caused by RNA polymerase
- Either strand of DNA can serve as the template.
- RNA Polymerase unwinds and rewinds the DNA, adding 5’ nucleoside triphosphates to the RNA’s 3’ hydroxyl.
- The template stand is read 3’ to 5’, and the direction of synthesis is 5’ to 3’.
- RNA Polymerase does possess some proofreading ability and has an error rate of about 1 in 10,000, while DNA Polymerase III has an error rate of about 1 in 10,000,000. (Please discuss.)
- It doesn’t matter that much. it’s one RNA molecule out of many many.
- If DNA get mutated, thats a big deal, however.
- Actinomycin D and Mitomycin intercalate between two G-C base pairs in the DNA and inhibit the RNA Polymerase thusly acting as antibiotic
- Rifampicin binds to the beta subunit of bacterial RNA Polymerase, and subsequently prevents the formation of the first phosphodiester bond.
- Used for treatment of tuberculosis; does NOT work on eukaryotes.
- Figure -
- Start with unwinding of the stands going from 5’ –>3’
- Once RNA is made, old RNA will start to dissociated from DNA and just dangle around

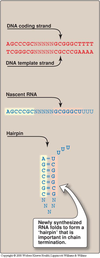
Termination of transcription
- Rho-independent termination of transcription
- Top: DNA template sequence generates a self-complementary sequence in the RNA
- Bottom: Hairpin structure formed by the RNA (N represents a non-complementary base)
- Signals termination
- Termination can be either associated with the rho factor or be independent of this factor.
- The rho –independent termination occurs as a result of the RNA’s forming a hairpin loop when it transcribes a palindromic sequence ( “A man, a plan, a canal, Panama“).
Rifampicin
- Rifampicin does NOT bind eukaryotic RNA Polymerase.
- Does NOT affect eukaryotes
- Prevent prokaryotic RNA polymerase activity.

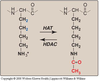
Histone
- Acetylation/deacetylation of a lysine residue in a histone
- Acetylation removes the positive charge; not going to be able to bind to DNA thus leading to more transcription as DNA strands are more available.
- HAT is histone acetyltranssferase;
- HDAC is histone deacetylase
- Exposure to BPA lead to hyper-acetylation leading to autism in child
Chromatin
- Euchromatin is the more relaxed form of chromatin, allowing a greater degree of transcription. Heterochromatin is the more condensed form, with a lesser amount of transcription, e.g. female barr body; inactivated X chromosome
- The acetylation of the lysine residues on histones by histone acetyltransferase (a) reduces the positive charge on the lysine and (b) decreases the interaction of the histone with the DNA (c) relaxing the chromatin.
- The acetyl group is removed by histone deacetylase, condensing the chromatin.
- This ongoing process is an integral part of histone remodeling.
Trans acting factors
- Trans acting factors
- There are three eukaryotic RNA polymerases:
- RNA Polymerase I…transcribes the 28S, 18S, and 5.8S rRNA genes in the nucleolus
- RNA Polymerase II…transcribes mRNA genes,snoRNA, snRNA genes (the only one we have to know)
- RNA Polymerase III…transcribes tRNA genes and 5S rRNA gene
Transcription factors
- There also are general transcription factors that bind to promoters. They include the following; TFIIA, TFIIB,…THIIH, also, TBP (TATA binding protein) and TBF’s (TBP Associated Factors). Usage of specific transcription factors depends on the specific gene and the specific situation.
- These factors are characterized as having structural motifs as leucine zippers, zinc fingers, helix-loop-helix, bromodomains, etc.
- Transcription factor work as combinatorial fashion; work together to induce specific transcription
- Promotor region is cis, whereas protein or other factors that bind to them are trans.
- Transcription fcators for prokaryotes = sigma factor (the only one)
- Eukaryotes are a lot more complicated and lot more varieties
- Cis acting factors are nucleotide sequences on the DNA, as opposed to trans acting (protein) factors that interact with these cis factors.
- cis-Regulatory Elements (CREs) are regions of non-coding DNA which regulate the transcription of nearby genes (wiki)
- The eukaryotic promoters contain the following conserved sequences:
- TATA (Hogness) box, found at -25
- CAAT box, found at -70
- GC boxes, found between -40 and -110
- These regions of the eukaryotic promoter bind the trans acting factors.
- Figure: Eukaryotic gene promoter consensus sequence

Transacting factors
- The information can arise from outside of the cell and be transmitted through signaling pathways.
- An example would be JAK-STAT ( Janus Kinase Signal Transducer and Activator of Transcription) [we don’t have to know this]. The cellular receptor is activated by a signal from different cytokines ( interferon, interleukin, etc.). Ultimately, the STAT protein translocates into the cell nucleus, binds to the promoter, and initiates the transcription of specific genes.
- Enhancers (or silencers) function in the stimulation or the inhibition of transcription
- They can be located either (a) 5’ or 3’ to the transcription start site, (b) close to or far away from the transcription start site (up to 50,000 base pairs away), (c) on either strand of the DNA.
- These enhancers contain response elements ( Hormone response element, Glucocorticoid response element, Cyclic AMP response element, etc.) that interact with specific trans acting factors.
- Transacting factors are activated by physiological change/message from somewhere in the body ; involve some genes to be transcribed, cascade of events, eventually leading to TF’s binding to DNA
- The effect of these enhancers/silencers is achieved by the looping out of the DNA between the enhancer and the promoter. This allows for contact between the transcription factors bound to the enhancer with those bound to the promoter.
- Figure:
- A: Assembly of eukaryotic initiation complex
- B: Enhancer stimulation of RNA polymerase II
- Transcription factosrs bind promotoer and amplify transcription
- Enhancers, after being binded to transcription factors, will affect TF’s that are very far downstream, which then either lead to enhancing or silencing effect.

Possible locations of enhancer sequences
- Possible locations of enhancer sequences

Posttranscriptional Modification of RNA
- Messenger RNA precursor molecules are called heterogeneous nuclear RNA (hnRNA)–>immature mRNA, which is sent to ribosomes
- 5’ “capping”: to allow translation beggins
- Poly-A tail: to help stabilize the mRNAs and facilitate their exit from the nucleous