Chapter 26: RNA Metabolism Flashcards
DNA-Dependent Synthesis of RNA
Transcription differs from replication in that it
- does not require a primer
- involves only limited segments of a DNA molecule
- only one DNA strand serves as a template for a particular RNA molecule
DNA-Dependent Synthesis of RNA
Template and nontemplate (coding) DNA strands
- template strand
- the strand that serves as template for RNA synthesis
- nontemplate strand, or coding strand
- DNA strand complementary to the template
- template DNA strand is copied in the 3’→5’ direction
- antiparallel to the new RNA strand
- Polymerase elongates RNA strand in 5’→3’ direction
- RNA transcript is identical in sequence (with U in place of T) to the nontemplate/coding strand
- regulatory sequences that control transcription are designated by the sequences in the coding strand
DNA-Dependent Synthesis of RNA
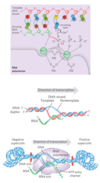
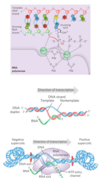
DNA-dependent RNA polymerase
- most active when bound to a double-stranded DNA
- requires DNA template, all four ribonucleoside 5’-triphosphates, Mg2+ and one Zn2+
- elongates RNA strand in 5’→3’ direction by adding ribonucleotide units to the 3’-hydroxyl end
- 3’-hydroxyl group acts as a nucleophile, attacking the α phosphate of the incoming ribonucleoside triphosphate and and releasing pyrophosphate
- template DNA strand is copied in the 3’→5’ direction
- antiparallel to the new RNA strand
- RNA transcript is synthesized on the template strand and is identical in sequence (with U in place of T) to the nontemplate/coding strand
Transcription
- Initiation occurs when RNA polymerase binds at a proromter (specific DNA sequences)
- The 5’-triphosphate group of the first residue in a nascent (newly formed) RNA molecule is not cleaved to release PPi
- reaction
- involves two Mg2+ ions and three Asp residues (highly conserved in RNA polymerases of all species)
- One Mg2+ ion facilitates attack by the 3’-hydroxyl group on the α phosphate of the NTP
- Second Mg2+ ion facilitates displacement of the pyrophosphate
- Both Mg2+ stabilize the pentacovalent transition state
- growing end of new RNA strand base-pairs temporarily with DNA template to form a short hybrid RNA-DNA double helix, 8 bp long
- About 17 bp of DNA are unwound at any given time
- DNA is unwound ahead and rewound behind as RNA is transcribed
- As the DNA is rewound, the RNA-DNA hybrid is displaced and the RNA strand is extruded and DNA duplex reforms
- Movement of an RNA polymerase along DNA creates positive supercoils (overwound DNA) ahead of the transcription bubble and negative supercoils (underwound DNA) behind it
- polymerase footprint encompasses about 35 bp of DNA during elongation
Inhibition
- actinomycin D and Acridine
- inhibits elongation in both bacteria and eukaryotes
- inserts (intercalates) into the doublehelical DNA between successive G☰C base pairs, deforming the DNA
- prevents movement of polymerase
- Rifampicin
- inhibits bacterial RNA synthesis
- binds to the β subunit of bacterial RNA polymerases
- prevents the promoter clearance
- α-amanitin
- disrupts mRNA formation
- blocks Pol II and, at higher concentrations, Pol III
DNA-dependent RNA polymerase
Bacteria
E. coli Structure
- a holoenzyme, large and complex
- five core subunits: α2ββ’ω; Mr 390,000
- transient 6th subunit: σ
- binds transiently to the core and directs the enzyme to specific binding sites on the DNA
- most common is σ70
- exists in several forms, depending on the type of σ subunit
- lacks a separate proofreading 3’→5’ exonuclease active site
- higher error rate for transcription than replication
- one error for every 104 to 105 ribonucleotide
- cuz many copies of an RNA are made and eventually degraded and replaced it is of less consequence than mistakes in DNA
- does pause when a mispaired base is added
- can remove mismatched nucleotides from 3’ end by direct reversal of polymerase reaction
- don’t know if this qualifies as proofreading function and to how much it contributes to fidelity of transcription
DNA-Dependent Synthesis of RNA
Initiation of RNA synthesis
Bacteria
E. coli Promoters
- RNA polymerase binds to promoters
- different types depending on σ factor used
- PDF pg. 1095, table 26-1 lists some
- using different σ subunits, allows cell ability to coordinate which genes to express
- availability of the various σ subunits depends on
- regulated rates of synthesis and degradation
- postsynthetic modifications that switch a σ subunit between active and inactive
- anti-σ proteins, each type binding to and sequestering a particular σ subunit
- σ70 promoter
- sequences can be variable
- binding occurs ≈ between -70 and -35
- 70 bp before transcription start site to 30 bp after it
- -10 and -35
- interaction sites for the σ70 subunit
- consensus sequence
- -10 is (5’)TATAAT(3’)
- -35 region is (5’)TTGACA(3’)
- UP (upstream promoter) element
- AT-rich recognition element
- between positions -40 and -60
- in promoters of certain highly expressed genes
- bound by the α subunit
- binding efficiency depends on promoter
- the consensus sequences it has
- spacing between them
- their distance from the transcription start site
- promoters establish a basal level of expression, varying from gene to gene
- σ32 promoter
- heat shock genes
- made at higher levels when the cell has received an insult
- PDF pg. 1092, Figure 26-5
Process
- σ factor binds to polymerase and directs it to the promoter
- polymerase binds to the promoter
- A closed complex where bound DNA is intact forms
- An open complex where the bound DNA is intact and partially unwound near the -10 sequence forms
- Transcription is initiated
- complex converts to the elongation form
- complex moves away from the promoter (promoter clearance)
- σ factor disassociates at random
- NusA binds to polymerase, where σ subunit binded
- When transcription finishes
- NusA dissociates polymerase
- polymerase dissociates from the DNA
- σ subunit can bind again to polymerase
DNA-Dependent Synthesis of RNA
Transcription Regulation
- can occur at any step in transcription
- Transcription is the first step in pathway of protein synthesis, so much of the regulation of protein levels in both bacterial and eukaryotic cells is directed at transcription, particularly its early stages
- binding of proteins to sequences both near to and distant from the promoter can also affect levels of gene expression
- cAMP receptor protein (CRP)
- increases the transcription of genes coding for enzymes that metabolize sugars other than glucose when cells are grown in the absence of glucose
- Repressors are proteins that block the synthesis of RNA at specific genes
DNA-Dependent Synthesis of RNA
Footprinting
- identifies the DNA sequences bound by a particular protein
- isolate a DNA fragment thought to contain sequences recognized by a DNA-binding protein
- radiolabel one end of one strand
- Tube 1
- same DNA sample
- Treat with DNase to introduce random breaks in the DNA fragment
- Tube 2
- same DNA sample
- Add DNA-binding protein
- Treat with DNase to introduce random breaks in the DNA fragment
- use electrophoresis to produce a ladder of radioactive bands for each tube
- compare the 2 gels side by side
- A gap in Tube 2 identifies the sequences that the protein binds
- DNase was unable to make any cuts here
- PDF pg 1093
DNA-Dependent Synthesis of RNA
Transcription
Bacteria
- RNA polymerase will introduce a large number of nucleotides into a growing RNA molecule before dissociating
- if an RNA polymerase released an RNA transcript prematurely, it could not resume synthesis of the same RNA, it would have to start again
DNA-Dependent Synthesis of RNA
Transcription Termination
Bacteria
- certain DNA sequences results in a pause in RNA synthesis, and some result in transcription termination
- least two classes of termination signals:
- ρ (Rho-dependent terminators)
- has a CA-rich sequence called a rut (rho utilization) element
- The rho protein associates with the RNA at specific binding sites and migrates in the 5’→3’ direction following the RNA polymerase
- has ATP-dependent RNA-DNA helicase activity that helps protein move along the RNA
- RNA polymerase pauses at a termination site
- rho catches up
- rho unwind the RNA–DNA in transcription bubble
- ATP is hydrolyzed by rho during termination process
- transcription stops, mRNA is released
- ρ-independent terminators
- have inverted repeats
- self-complementary sequences
- can form a hairpin structure
- 15 to 20 nucleotides before end of RNA strand
- followed by three A residues
- highly conserved
- transcribed into U residues
- polymerase reaches terminator with this region
- inverted repeats are transcribed into RNA
- string of Us causes the polymerase to pause
- inverted repeats in RNA fold into a hairpin
- hairpin disrupts several A═U base pairs in the RNA-DNA hybrid
- may disrupt important interactions between RNA and polymerase, facilitating dissociation of the transcript
- have inverted repeats
- ρ (Rho-dependent terminators)
DNA-Dependent Synthesis of RNA
Eukaryotes: RNA Polymerases
RNA polymerase I
- synthesis of preribosomal RNA (pre-rRNA)
- contains the precursor 18S, 5.8S and 28S rRNAs
- promoters differ greatly in sequence from one species to another
RNA polymerase III
- makes tRNAs, 5S rRNA, other small specialized RNAs
- some of the sequences required for the regulated initiation of transcription by Pol III are located within the gene itself
RNA polymerase II
- synthesis of mRNAs and some specialized RNAs
- recognize thousands of promoters
- Some Pol II promoters have a few sequence features in common
- TATA box: TATA(A/T)A(A/T) (A/G) near base pair -30
- Inr sequence (initiator) near the RNA start site at 11
- The DNA is unwound at the initiator sequence (Inr)
- transcription start site is usually within or very near this sequence
- not too many of these
- Pol II operates at many promoters that lack these features
- Struture
- huge enzyme with 12 subunits
- RBP1
- largest subunit
- homologous to β subunit of bacterial
- has a long carboxyl-terminal domain (tail) (CTD)
- has many repeats of a consensus heptad amino acid sequence –YSPTSPS–
- separated from the main body of the enzyme by linker sequence
- many roles
- RBP2 is structurally similar to the bacterial β subunit
- RBP3 and RBP11 similar to the bacterial α subunits
- Requires general transcription factors to form the complex
- TFII with an additional identifier
- highly conserved
- Assembly
- Promoter with TATA box
- TATA-binding protein (TBP) binds to the TATA box
- Promoter w/out TATA box
- TBP arrives as part of a multisubunit complex called TFIID
- closed complex is formed
- TFIIB binds TBP and DNA on both sides of TBP
- TFIIA binds
- TFIIA and TFIIB stabilize the TBP-DNA complex
- TFIIF and Pol II binds TFIIB-TBP
- TFIIF
- helps target Pol II to its promoters
- reducies the binding of the polymerase to nonspecific sites on the DNA
- TFIIE and TFIIH bind to create the closed complex
- TFIIH
- has DNA helicase activity
- unwinds DNA near the start site
- creates open complex
- essential in the repair of damaged DNA
- repair is more efficient within genes that are actively being transcribed
- template strand is repaired more efficiently than the nontemplate strand
- can interact with the lesion and recruit the entire nucleotide-excision repair complex
- Promoter with TATA box
- Strand Initiation and Promoter Clearance
- TFIIH has a kinase activity and phosphorylates Pol II at many places in the CTD
- Other protein kinases (CDK9) also phosphorylate the CTD on the Ser residues
- Phosphorylation causes a conformational change, which intitiate transcription
- Phosphorylation state of the CTD changes as transcription proceeds
- affect the interactions between the transcription complex and other proteins and enzymes
- TFIIE and then TFIIH is released during synthesis of the initial 60 to 70 nucleotides
- Pol II enters the elongation phase of transcription
- Elongation, Termination and Release
- TFIIF
- remains associated with Pol II throughout elongation
- enhances the activity of the polymerase
- some bind to CTD
- suppress pausing during transcription
- Once the RNA transcript is completed, transcription is terminated
- Pol II is dephosphorylated and recycled
- TFIIF
RNA Processing
- newly synthesized RNA molecule is called a primary transcript
- introns are noncoding tracts that break up the coding region of the transcript
- exons are coding segments
- introns are removed and exons are joined to form a continuous sequence
- 5’ cap is added at the 5’ end
- 3’ end is cleaved and 80 to 250 A residues are added to create a poly(A) “tail.”
- protein complexes that carry out these three mRNA-processing reactions
- do not operate independently
- organized in association with each other and with the phosphorylated CTD of Pol II
- affects the function of the others
- the primary transcript is synthesized, ensconced in an elaborate complex involving dozens of proteins
- The composition of the complex changes as the primary transcript is processed, transported to the cytoplasm, and delivered to the ribosome
5’ cap
- in most eukaryotic mRNAs
- a residue of 7-methylguanosine linked to the 5’-terminal residue of the mRNA via a 5’,5’-triphosphate linkage
- helps protect mRNA from ribonucleases
- binds to a specific cap-binding complex
- participates in binding of the mRNA to the ribosome
- Synthesis of the cap
- carried out by enzymes tethered to the CTD of Pol II
- formed by condensation of a molecule of GTP with the triphosphate at the 5’ end of the transcript
- guanine is methylated at N-7,
- additional methyl groups are often added at the 2’ hydroxyls of the first and second nucleotides adjacent to the cap
- occurs very early in transcription, after the first 20 to 30 nucleotides of the transcript have been added.
- the three capping enzymes have the 5’ running through them and are linked with the RNA polymerase II CTD until the cap is synthesized
- The capped 5’ end is then released from the capping enzymes and bound by the cap-binding complex
Splicing Introns
- vast majority of genes in vertebrates contain introns; among the few exceptions are those that encode histones
- found in a few bacterial and archaeal genes
- In eukaryotic mRNAs, most exons are less than 1,000 nucleotides long
- 100 to 200 nucleotide size range
- encoding stretches of 30 to 60 amino acids within a longer polypeptide
- Introns vary in size from 50 to 20,000 nucleotides
- higher eukaryotes have much more introns than to exons
- 4 types of introns
Group I & Group II
- self-splicing
- doesn’t require a high-energy cofactor (such as ATP)
- involve two transesterification reaction steps
- a ribose 2’- or 3’-hydroxyl group makes a nucleophilic attack on a phosphorus and a new phosphodiester bond is formed at the expense of the old, maintaining the balance of energy
- reactions similar to DNA breaking & rejoining reactions promoted by topoisomerases and site-specific recombinases
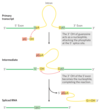
- Group I
- found in some nuclear mitochondrial, and chloroplast genes that code for rRNAs, mRNAs, and tRNAs, and bacteria
- requires a guanine nucleoside or nucleotide cofactor
- 3’-hydroxyl group of guanosine is the nucleophile in the first step
- it forms a normal 3’,5’-phosphodiester bond with the 5’ end of the intron
- 3’ hydroxyl of the exon that is displaced then acts as a nucleophile at the 3’ end of the intron
- result in excision of the intron and ligation of the exons
- Group II
- found in primary transcripts of mitochondrial or chloroplast mRNAs in fungi, algae, plants, and bacteria
- reaction pattern is similar to Group I
- 2’-hydroxyl group of an A residue within the intron is the nucleophile
- a branched lariat structure is formed as an intermediate
- spliced intron is eventually degraded
spliceosomal introns
- nuclear mRNA primary transcript introns
- third and largest class of introns
- not self-splicing
- undergo splicing by the same lariat-forming mechanism as the group II introns
- have dinucleotide sequence GU at the 5’ end and AG at the 3’ end
- mark the sites where splicing occurs
- spliceosomes (U1 and U2) removes introns
- made up of specialized RNA protein complexes, small nuclear ribonucleoproteins (snRNPs)
- Each snRNP contains one small nuclear RNAs (snRNAs)
- a class of eukaryotic RNAs, 100 to 200 nucleotides long
- Five snRNAs (U1, U2, U4, U5, and U6)
- found in abundance in eukaryotic nuclei
- highly conserved in eukaryotes
- snRNPs together contribute five RNAs and about 50 proteins to the core spliceosome
- snRNPs together contribute five RNAs and about 50 proteins to the core spliceosome having multiple functions: in splicing, mRNA transport to the cytoplasm, translation, and eventual mRNA degradation
- ATP is required for assembly of the spliceosome
- Some components of the splicing apparatus are tethered to the CTD of RNA polymerase II
- spliceosomes (U11 and U12)
- less common type of spliceosome
- U1 and U2 snRNPs are replaced by the U11 and U12 snRNPs
- remove a rare class of introns that have (5’)AU and AC(3’) terminal sequences
- Splicing pathway
- U1 snRNA contains a sequence complementary to sequences near the 5’ splice of spliceosomal introns
- U1 snRNP binds to this region
- U2 is paired to the intron at a position encompassing the A residue that becomes the nucleophile during the splicing reaction
- U2 pairing causes a bulge
- displaces and helps to activate the adenylate, the 2’ OH which will form the lariat structure through a 2’,5’-phosphodiester bond
- U4-U6 bind to form an inactive spliceosome
- Internal rearrangements expel U1 and U4 and activate spliceosome
- U6 is paired with ‘5’ splice site and U2
- catalytic steps, which parallel those of the splicing of group II introns follow
- Coordination of splicing and transcription brings the two splice sites together
- spliced intron is eventually degraded
- PDF PG. 1105
Intron Group I Splicing
Intron Group II Splicing
3’ end, polyA tail
- have a string of 80 to 250 A residues
- binding site for one or more specific proteins
- probably help protect mRNA from enzymatic destruction
- Many bacterial mRNAs also acquire poly(A) tails, but these tails stimulate decay of mRNA
- Addition Pathway
- Pol II synthesizes RNA beyond the segment of the transcript containing the cleavage signal sequences, where the poly(A) tail is to be added, past the highly conserved upstream sequence (5’)AAUAAA
- RNA is cleaved by the endonuclease at a point 10 to 30 nucleotides 3’ to (downstream of) the sequence AAUAAA
- by a large enzyme complex with the CTD of RNA polymerase II
- Cleavage generates the free 3’-hydroxyl group defining the end of the mRNA
- A residues are immediately added by polyadenylate polymerase
- does not require a template
- requires the cleaved mRNA as a primer
- synthesizes a poly(A) tail 80 to 250 nucleotides
- If there are two or more sites for cleavage and polyadenylation, use of the one closest to the 5’ end will remove more of the primary transcript sequence, this is called polyA site choice

Differential RNA Processing
- Posttranscriptional processing is not limited to mRNA
- Ribosomal RNAs of bacterial, archaeal, and eukaryotic cells are made from longer precursors called preribosomal RNAs, or pre-rRNAs
- Transfer RNAs are similarly derived from longer precursors. These RNAs may also contain a variety of modified nucleosides
Ribosomal RNAs
Bacterial
- 30S RNA
- precursor for 16S, 23S, and 5S rRNAs
- and some tRNAs, although most tRNAs are encoded elsewhere
- about 6,500 nucleotides
- Before cleavage, the 30S RNA precursor is methylated at specific bases
- some uridine residues are converted to pseudouridine or dihydrouridine residues
- some occur on bases and some on 29-hydroxyl groups
- 16S rRNA has 11 modifications: one a pseudouridine and 10 nucleosides methylated on the base or the 2’-hydroxyl group or both
- 23S rRNA has 10 pseudouridines, 1 dihydrouridine, and 12 methylated nucleosides
- each modificaiton catalyzed by a different enzyme
- Methylation reactions use S-adenosylmethionine as cofactor
- No cofactor is required for pseudouridine formation
- RNA at both ends of the 30S precursor and segments between the rRNAs are removed during processing
- Coding sequences for tRNAs are also found on the 3’ side of the 5S rRNA in some precursor transcripts
- Cleavage liberates precursors of rRNAs and tRNA(s).
- Cleavage at points 1, 2, and 3 is carried out by the enzymes RNase III, RNase P (a ribozyme), and RNase E, respectively
- The final 16S, 23S, and 5S rRNA products result from the action of a variety of specific nucleases
- The seven copies of the gene for pre-rRNA in the E. coli chromosome differ in the number, location, and identity of tRNAs included in the primary transcript.
- Some copies of the gene have additional tRNA gene segments between the 16S and 23S rRNA segments and at the far 39 end of the primary transcript

Ribosomal RNAs
Eukaryotes
- process is initiated in the nucleolus, in large complexes that assemble on the rRNA precursor as it is synthesized by Pol I
- During transcription, the 45S primary transcript is incorporated into a nucleolar 90S preribosomal complex
- rRNA processing and ribosome assembly are tightly coupled here
- 45S precursor is processed
- methylated at more than 100 of its 14,000 nucleotides on the bases or the 2’-OH groups
- most common nucleoside modifications are conversion of uridine to pseudouridine and adoMet-dependent nucleoside methylation (often at 2’-hydroxyl groups)
- rely on snoRNA-protein complexes, or snoRNPs, each consisting of a snoRNA and four or five proteins
- snoRNPs
- box H/ACA snoRNPs are involved in pseudouridylylation
- box C/D snoRNPs function in 2’-O-methylations
- same enzyme may participate in modifications at many sites, guided by the snoRNAs
- snoRNAs
- 60 to 300 nucleotides long
- encoded within the introns of other genes and cotranscribed with those genes
- includes a 10 to 21 nucleotide sequence complementary to a site on rRNA
- has conserved sequence elements in the remainder of the snoRNA
- fold into structures bound by the snoRNP proteins
- snoRNPs
- conversion to dihydrouridine also occur
- A series of enzymatic cleavages of the 45S precursor produces the 18S, 5.8S, and 28S rRNAs
- mediated by endo- or exoribonucleases and nucleoside modification reactions
- requires small nucleolar RNAs (snoRNAs) found in protein complexes (snoRNPs) in the nucleolus that are reminiscent of spliceosomes
- guide nucleoside modification and some cleavage reactions, and ribosomal proteins
- Some pre-rRNAs also include introns that must be spliced
- these ribosomal subunits take shape with the assembling ribosomal proteins
- a tight coupling between rRNA transcription, rRNA maturation, and ribosome assembly
- The 5S rRNA is produced separately by a different polymerase (Pol III)
- PDF PG 1110, FIGURE 26–24 Processing of pre-rRNA transcripts
- PDF PG 1111, FIGURE 26–25 Functions of snoRNAs in guiding rRNA modification
Transfer RNAs
Transfer RNAs
- 40 to 50 distinct tRNAs
- eukaryotic cells have multiple copies of many of the tRNA genes
- derived from longer RNA precursors by enzymatic removal of nucleotides from the 5’ and 3’ ends
- In eukaryotes, introns are present
- 5’ end is processed first
- endonuclease RNase P
- removes RNA at the 5’
- contains both protein and RNA
- in bacterial cells it can carry out its processing function without the protein component
- example of a catalytic RNA
- endonuclease RNase P
- 3’ end is processed second
- 3’ end of tRNAs is processed by one or more nucleases including the exonuclease RNase D
- 3’-terminal trinucleotide CCA(3’) to which an amino acid is attached is absent and added during processing
- carried out by tRNA nucleotidyltransferase
- binds three ribonucleoside triphosphate precursors in separate active sites
- catalyzes formation of the phosphodiester bonds to produce the CCA(3’) sequence
- not dependent on a DNA or RNA template
- template is the binding site of the enzyme
- While the ends are being processed, specific bases in the rest of the transcript are modified
- modification of some bases by methylation, deamination, or reduction
- In the case of pseudouridine, the base (uracil) is removed and reattached to the sugar through C-5

Special-Function RNAs
snRNAs and snoRNAs
- facilitate RNA processing reactions
- synthesized as larger precursors and then processed
- snoRNAs
- many encoded within the introns
- introns are spliced from the pre-mRNA
- snoRNP proteins bind to the snoRNA
- ribonucleases remove the extra RNA at the 5’ and 3’ ends
- snRNAs destined for spliceosomes
- synthesized as pre-snRNAs by RNA polymerase II
- ribonucleases remove the extra RNA at each end
- nucleosides in snRNAs are also subject to 11 types of modification, with 2’-O-methylation and conversion of uridine to pseudouridine predominating
MicroRNAs (miRNAs)
- involved in gene regulation
- noncoding RNAs
- about 22 nucleotides long
- complementary in sequence to particular regions of mRNAs
- regulate mRNA function by cleaving the mRNA or suppressing its translation
- found in multicellular eukaryotes
- may target up to one-third of human mRNAs.
- synthesized from much larger precursors, in several steps
- primary transcripts for miRNAs (pri-miRNAs) vary greatly in size
- some are encoded in the introns of other genes
- PDF PG. 1112, FIGURE 26–27 Synthesis and processing of miRNAs
RNA Enzymes
- best-characterized ribozymes are the self-splicing group I introns, RNase P, and the hammerhead ribozyme
- two fundamental reactions: transesterification and phosphodiester bond hydrolysis (cleavage)
- substrate is often an RNA molecule, may be part of the ribozyme itself
- When substrate is RNA, the RNA catalyst makes use of base-pairing interactions to align the substrate for the reaction
- vary in size
- inactivated by
- heating above melting temperature
- addition of denaturing agents
- addition of complementary oligonucleotides
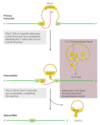
Group I Introns
- self splicing
- Binding of guanosine cofactor to the Tetrahymena group I rRNA intron is saturable (Km ≈ 30 μM) and can be competitively inhibited by 3’-deoxyguanosine
- internal guide sequence
- results in precise excision reaction
- can base-pair with exon sequences near 5’ splice site
- pairing promotes alignment of specific bonds to be cleaved and rejoined
- intron itself is chemically altered during the splicing reaction—its ends are cleaved
- synthesis
- after excision, the 414 nucleotide intron from Tetrahymena rRNA can, in vitro, act as a true enzyme
- cleavage reactions in the excised intron leads to the loss of 19 nucleotides from its 5’ end
- A G residue (shaded light red in figure) is added at the 5’
- was part of the removed sequence
- A portion of the internal guide sequence remains at the 5’ end of L-19 IVS
- remaining 395 nucleotide—referred to as L-19 IVS (intervening sequence)
- linear RNA
- promotes nucleotidyl transfer reactions where oligonucleotides are lengthened at the expense of others
- enzymatic activity of L-19 IVS ribozyme
- a cycle of transesterification reactions mechanistically similar to self-splicing
- 3’ OH of the G residue at the 3’ end of L-19 IVS plays a key role in this cycle
- this is not the G residue added in the splicing reaction
- (C)5 is one of the ribozyme’s better substrates because it can base-pair with the guide sequence remaining in the intron
- can process ≈ 100 substrate molecules per hour
- not altered in the reaction
- intron acts as a catalyst
- follows Michaelis-Menten kinetics
- specific for RNA oligonucleotide substrates
- can be competitively inhibited
- The kcat/Km (specificity constant) is 103
m-1s-1, lower than that of many enzymes - accelerates hydrolysis by a factor of 1010 relative to the uncatalyzed reaction
- makes use of substrate orientation, covalent catalysis, and metal-ion catalysis— strategies used by protein enzymes.
Other Ribozymes
- RNase P
- recognizes the 3D shape of pre-tRNA substrate, along with the CCA sequence, and cleaves the 5’ leaders from diverse tRNAs
- has an RNA component (the M1 RNA, with 377 nucleotides) and a protein component (Mr 17,500)
- under some conditions, the M1 RNA alone is capable of catalysis cleaving tRNA precursors
- protein stabilizes the RNA or facilitate its function in vivo
- virusoids
- small RNAs associated with plant RNA viruses
- self-cleavage reaction
- hammerhead ribozyme is an example
PDF pg. 115
mRNA Degradation
- Degradative pathways ensure that mRNAs do not build up in the cell and direct the synthesis of unnecessary proteins
- rates of degradation vary greatly for mRNAs
- Gene products needed constantly by the cell may have mRNAs that are stable over many cell generations
- RNA is degraded by ribonucleases
- In bacteria
- average half-life of the mRNAs of a vertebrate cell is about 3 hours
- mRNA turning over about 10 times per cell generation
- process begins with one or several cuts by an endoribonuclease, followed by 3’→5’ degradation by exoribonucleases
- hairpin structure in bacterial mRNAs with a ρ-independent terminator confers stability against degradation
- Similar hairpin structures can make some parts of a primary transcript more stable, leading to nonuniform degradation of transcripts
- In eukaryotes
- half-life of bacterial mRNAs is much shorter—only about 1.5 min
- have an exosome
- complex of up to 10 conserved 3’→5’ exoribonucleases
- involved in the processing of the 39 end
- both the 3’ poly(A) tail and the 5’ cap are important to the stability of many mRNAs.
- lower eukaryotes
- shortening the poly(A) tail
- decapping the 5’ end
- degrading the mRNA in the 5’→3’ direction
- higher eukaryotes
- also has a 3’→5’ degradative pathway which may be the major path




