Chromatin structure Flashcards
(65 cards)
What are topology associated domains (TADs)?
Chromatin interacting regions partitioned by sharp boundaries.
What stabilizes transcriptionally silenced regions?
ncRNA stabilizes transcriptionally silenced regions.
How do SWI-SNF and ISW1a complexes affect chromatin?
They alter nucleosome structure.
What is the structure of a nucleosome?
Nucleosome is composed of histones.
What is the role of histone post-translational modifications?
They impact chromatin structure.
What is the difference between euchromatin and heterochromatin?
Euchromatin is less condensed; heterochromatin is more condensed.
What happens during chromatin remodelling?
DNA is unwrapped and re-wrapped around nucleosomes.
What is the role of activation factors in gene expression?
They bind to gene promoters to activate genes.
How does estrogen affect gene transcription in different cells?
It transcribes ovalbumin in oviduct cells and vitellogenin in liver cells.
What controls transcription of numerous genes in bacteria?
A single repressor, as in the lac operon.
<p>why have genes constitituively inactive without the need for a repressor</p>
<p>More Economical</p>
What is required for gene activation in eukaryotes?
An activating factor binding to the gene promoter.
How can an activation factor be regulated?
It can be tissue-specific or inactivated by another binding factor.
<p>co-factor and activation factor process</p>
<p>In inactive tissue: Gene completely inactive and activating factor absent in inactive tissue</p>
<p>In Active tissue: Activation factor only synthesized in active tissue or active factor is also present in inactive tissue but requires a co - factor to stimulate gene expression.</p>
<img></img>
What could the co-factor for gene expression be?
A hormone or steroid.
<p>co-factor and cell type</p>
Expression of a different gene depending on the cell type.
What is an example of a co-factor affecting gene expression?
Estrogen treatment results in transcription of the ovalbumin gene.
What ensures cells remain in a differentiated state?
Stable mechanisms maintain differentiated characteristics long-term, even without phenotypic traits.
Do cells of one type differentiate into different types over time?
No, they continue to differentiate into the same type throughout their lifetime.
What happens to cartilage cells reprogrammed to an undifferentiated state?
They can differentiate back into cartilage cells even after 20 passages.
What maintains the semi-stability of a cell’s differentiation state?
Patterns of association between DNA and proteins, i.e., chromatin structure.
<p>Nucleosome structre</p>
<p>H1</p>
<p>H2A</p>
<p>H2B.</p>
<p>H3</p>
<p>H4</p>
<img></img>
<p>ACtive DNA structure</p>
<p>Beads on a string</p>
<p>Inactive DNA structure</p>
<p>30nmm chromatin fibre</p>
<img></img>
Linker region variance
Linker region can be shorter or longer ◦ Depends on presence (a) or absence (b) of Histone H1
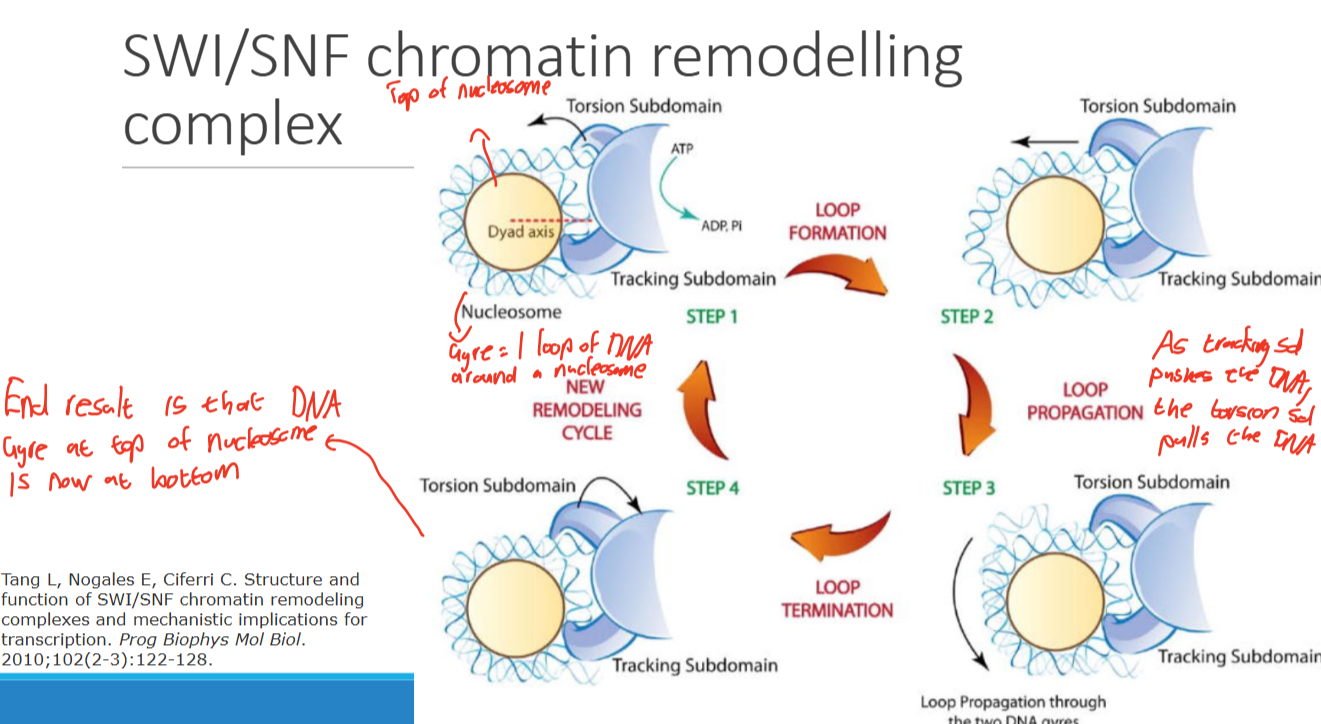
SWI/SNF chromatin remodeling complex mechanism
-Loop formation
-Loop propagation
_loop termmination
-New remodeling complex
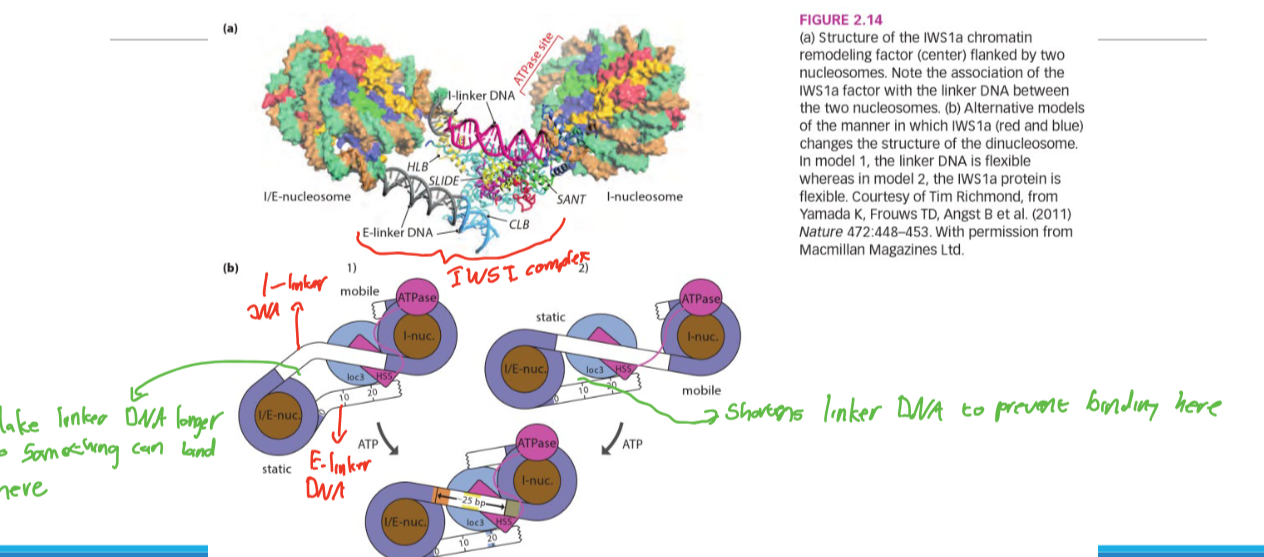
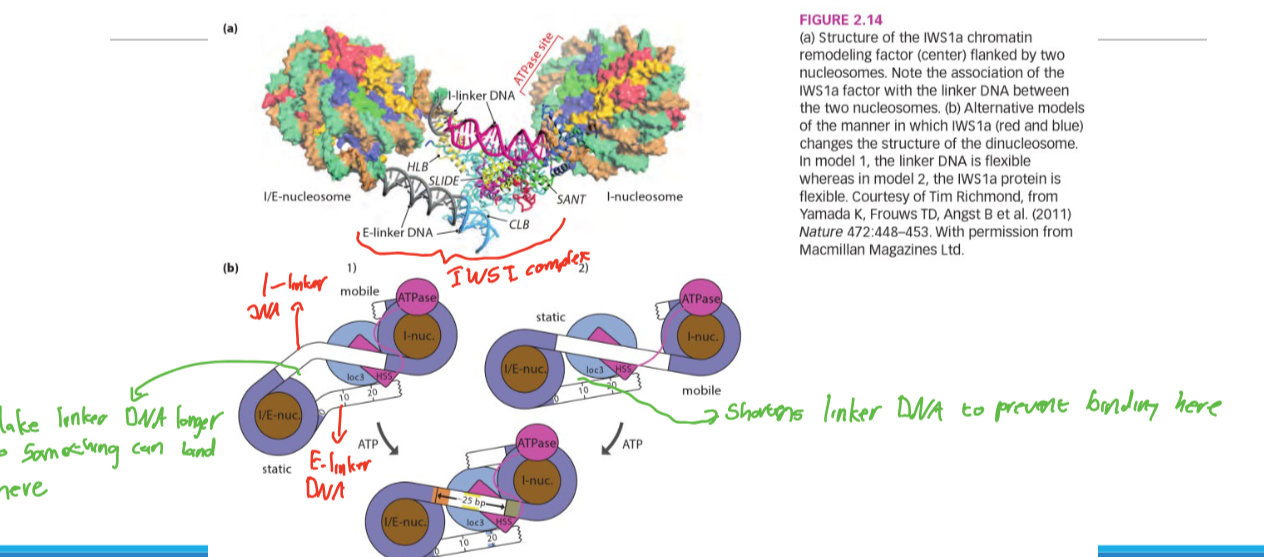
IWSI1 mechanism
Changes the distance between nucleosomes by changing linker DNA length
-This is doen to expose or hide certain regions of DNA iin order to change expression
How do chromatin remodeling complexes change nucleosome position
A) Nucleosome structure changes (changes how tightly its wrapped)– DNA exposed B) Nucleosome displacement ( changes the position of DNA on nnucleosome) – sliding C) Nucleosome displacement – nucleosome loss
Post - translational modifications ◦ Target amino terminal portion of histones ◦ Histone tails – beyond interior of nucleosome o Acetylation o Methylation o Ubiquitination and sumoylation o Phosphorylation
Three major families-
Gcn5 N - acetyl transferases (GNATs) ◦ P300/CBP family ◦ Morf , Ybf2, Sas2 and Tip60 (MYST) family
Acetyl groups are negative and reduce the postive charge of the lysine on histone tails- makes SNA more accesible by loosening chromatin structure
Methyl groups result in a neutral charge.
Examples of competitiveness between acetylation and methylation
Methylation of H3K9 prevents acetylation on itself as well as other sites as it all blocks H3K14 acetylation
No, its a competitive process where the tail can either be methylated or acetylated
Function of DNA phosphorylation
-Reduces affinity to DNA by reducing net positive charge
-It recruits other regulatory proteins and chromatin remodelling complexes
Histone code
Pattern of post-translational modifications
-Recognised by regulatory proteins
-Order of specific functional groups (phosphorylation and acetylation) on tail creates a specific binding site that directly fit into regulatory proteins
Ubiquitination
If it occurs on H2B then it promotes methylation of H3 at position 4 and 79
Small ubiquitin related modifier
SUMO recruits histone deacetylases.
-H3K16 interacts with a 7 aa cluster on H2A/H2B on neighboring nucleosome
-It reduces the positive charge which weakens H2A/H2B interaction on neighboring nucleosome.
-Promotes open chromatin structure.
Binds linker DNA,
seals 2 turns of DNA around core histone octamer
Can recruit DNA methyl transferases ( Dnmt ) that methylate DNA Associated with closed chromatin conformation Architectural protein – regulates chromatin structure
Allow for open chromatin structure in that region to increase gene expression
Regulate chromatin structure of large DNA regions.
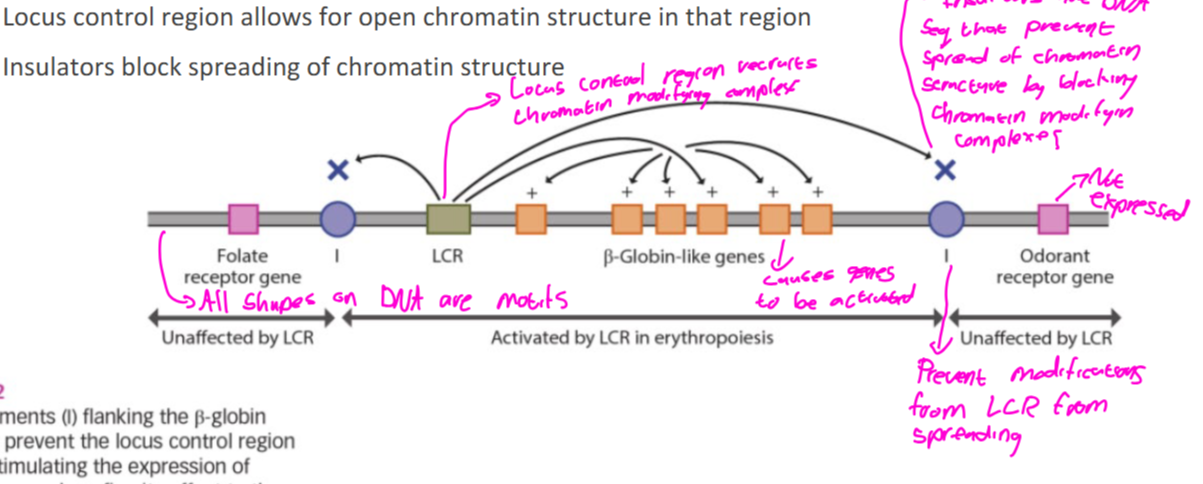
Bind insulators and form dimers
-The dimer has been associated with chromatin loops and may be associated with multiple insulator elemnets
-Loss of LCR and insulator regions can lead to improper gene regulation and disease
Centromeric protein A mechanism
Incorporated centromeric regions and binds to stellite DNA that is rich in A and T and contains a specific motif that allows CENPa to bind
-This causes a structural change that allows DNA to bend more
H2AX mechanism
When DNA has a double stranded break, H2AX replaces H2A
-This contains an extra serine residue that can be phosphorylated which signals DNA repair enzymes to activate
why is a double break bad?
It can cause chromosomal rearrangements which can cause disease
where is H1 found
Enriched in transcriptionally inactive regions Depleted in regions actively transcribed
how to determine looping patterns
Use chromatin conformation capture
NB of loops
-Can alter dtructure to beads to allow transcription
-Specific domains of chromatin allow for structure to be altered in a specific way to allow for transcription
Chromatin Conformation capture
Cell Fixation: Cells are treated with a cross-linking agent, such as formaldehyde, to covalently bond DNA to proteins and other nearby DNA regions, preserving their spatial arrangement.
Digestion: The fixed chromatin is then digested with a restriction enzyme, which cuts the DNA at specific sequences, leaving the cross-linked regions intact.
Ligation: The digested fragments are diluted to favor intramolecular ligation, where DNA ends that are physically close (looped) are joined together by DNA ligase. This step captures the spatial proximity of DNA regions.
Cross-link Reversal: The cross-links between DNA and proteins are reversed, typically by heating, to release the DNA from the proteins, leaving only the ligated DNA fragments.
Analysis: The ligated DNA is purified and analyzed, often using techniques like PCR, sequencing, or microarrays, to identify and quantify the interactions between different genomic regions.
-Spool of active DNA is left


