Exam 3: Molecular Genetics And Biotechnology Flashcards
(46 cards)
Describe Recombinant DNA Technology
You combine DNA from multiple sources, most often by isolating the desired stretches of DNA from the source and inserting them into a vector, which is a circular piece of DNA that can be cut open at one site and have a foreign DNA fragment ligated into it
How do you isolate your gene of interest using Restriction endnucleases?
You can use restriction endonucleases (aka restriction enzymes) to cut the desired fragments of DNA
Then you use endonucleases that leave single-stranded overhangs—i.e. “sticky ends”—to incorporate these DNA fragments into a vector

Isolating Your DNA Of Interest
Describe Briefly The Polymerase Chain Reaction (PCR)
and the ingredients
Another approach uses the polymerase chain reaction (PCR), which can make millions of copies of any stretch of DNA the researcher desires
For a PCR you need:
Template DNA—Source DNA to be copied
Deoxynucleotides in triphosphate form–dNTPs
Primers—single-stranded DNA ~20-25 nucleotides long
DNA polymerase
A buffer to control pH, [Mg+2] and other ions
Describe PCR in Detail
Heat the mixture to temperatures that denature the template DNA to make your template and primers single-stranded and ready for complementary sequences to bind
Heat the mixture to ~72oC, the temperature at which the DNA polymerase works fastest—the DNA polymerase reads the DNA and chains the complementary nucleotides together

How do you Screen For Vectors That Have Acquired The Desired Insert?
You often put your vector into a bacterium by transformation—ability of a bacterium to take DNA fragments or plasmids up from the environment
By a combination of calcium/heat/electrical stimulation, you render the bacterial cells competent—able to take up a plasmid from the environment
The process is inefficient—you end up with three types of bacteria:
Those that have not taken up a plasmid
Those that have taken up a plasmid that has no vector
Those that have taken up a plasmid that contains the vector
What does a typical vector contain?
An origin of replication
Enzyme cut sites
Genes that enable you to identify cells that have taken up vectors that have the desired insert in them
How do you screen bacteria to detect sucessful transformations?
The antibiotic ampicillin has a beta-lactam ring in its chemical structure
You put penicillin in the growth medium, so only cells that have the vector in them can grow in it
The lac Z gene enables you to identify vectors that have taken up the insert
Lac Z encodes beta-galactosidase, which converts an artificial substrate called X-gal (which is put on the plate) to a blue metabolite, creating blue colonies
Putting an insert in the vector disrupts the function of the lacZ gene, producing white colonies

Explain How You Determine The Size Of Your DNA Fragments
Gel electrophoresis involves running an electric current through a gel-like substance (agarose, acrylamide) and seeing how quickly the DNA fragment gets pushed through the cell by the electric current
Many gels separate fragments according to size—smaller ones migrate more quickly
How do you view DNA fragments?
The outside lanes of this gel are loaded with a size ladder—a collection of fragments of known size
This enables you to determine the size of your fragments
Of the Jones’ five children, did any of them inherit Mr. Jones’ deletion? (the 1 in the middle)

How Does Southern Blotting Allow You To Detect Specific Sequences In Genomic Digests
The genomic DNA of your organism of interest is digested by endonucleases and subjected to gel electrophoresis
After gel electrophoresis the fragments are denatured and transferred to a nylon membrane, which is incubated with a denatured and radioactive probe
The probe binds to complementary sequences in the DNA fragments
The membrane is placed against X-ray film and dark bands develop where probe has bound
Your choice of probe determines which fragments you will see

How do you create Genomic and cDNA Libraries?
You can isolate the mRNA from a tissue, use reverse transcription to create cDNAs from the mRNAs, clone the cDNAs into vectors and transform bacteria with the vectors, thereby creating a library of bacterial clones that together represent the set of genes that are active in that tissue under those circumstances
You can screen the bacterial colonies with the probe of your choice—the probe will hybridize to complementary sequences, thereby showing you which bacterial colonies contain that sequence

Name Some Applications That Capitalize On The Hybridization Between Complementary sequences
Nucleic acid fragments that have complementary sequences will bind to each other–If you want to see if a particular sequence is present, you can use a fragment of DNA or a cDNA that has a complementary sequence as a probe, as in Southern blotting
Hybridization is the basis for microarray-based assays, where a gene chip is spotted with thousands of different probes
In situ hybridization involves applying your probe to tissues, embryos, chromosomes—i.e. the natural site
In situ hybridization studies can detect gene expression, or deletions and duplications in chromosomes
Describe Situ Hybridization
FISH—Fluorecence in situ hybridization
Green probe binds to chromosome 1q telomere—identifies the two chromosome 1s
Red probe from 1p telomere reveals a deletion on one chromosome 1—the red probe did not find a complementary sequence
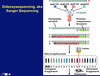
What is Dideoxysequencing, aka Sanger Sequencing?
A small portion of each nucleotide is present as a fluorescently labeled dideoxynucleotide—lacking the O on both the 2’ and the 3’ carbons
When a dideoxynucleotide gets incorporated into a newly synthesized DNA strand, the strand cannot be extended beyond that point, because there is no 3’ O to bind a new nucleotide to
Each of the four different dideoxynucleotides is labeled with a different fluorescent molecule, so each fragment that gets made by the PCR will fluoresce the color that corresponds to the last base in that fragment
Describe “Next Generation” sequencing- pyrosequencing?
Next generation sequencing sequences millions of fragments at the same time
Each fragment is attached to a bead and placed into a separate well in a tray that has millions of wells, the same way a microarray aligns millions of probes on a slide
The tray is incubated with a sequencing reaction mixture that contains only one type of deoxynucleotide triphosphate (ex. dATP)
If the A is going to be incorporated into the sequence, the two terminal phosphates are cleaved off (as one unit—a pyrophosphate molecule) to provide energy for the reaction
Describe the second part of “Next Generation” sequencing?
The tray is then flushed and incubated with a sequencing reaction mixture that contains another type of deoxynucleotide triphosphate (ex. dCTP)
If the C is going to be incorporated into the sequence, the two terminal phosphates are cleaved off (as one unit—a pyrophosphate molecule) to provide energy for the reaction
The sequencer detects this as a flash of light in that well
The machine cycles through exposing the plate to dA, dC, dG and dT numerous times
For each reaction mix (containing A, C, G or T), the sequencer records which wells showed the flash of light
The sequencer can assemble all this info to tell what the sequence of the fragment in each well is
What is Illumina Sequencing in “Next Genration” Sequencing?
The sequencing reaction is a lot like dideoxysequencing—different colored fluors are attached to the four different nucleotides, and each nucleotide has a molecule bound to it that will prevent the chain from being extended past that nucleotide
When a nucleotide analog gets incorporated into the newly synthesized strand, the chain cannot be extended farther
After the nucleotide is incorporated, the terminator and the fluor are taken off the nucleotide chemically
This allows the machine to detect which color fluor has been liberated, thereby telling it which nucleotide was incorporated into that spot, and
another nucleotide to be added onto the newly synthesized DNA chain
What is “Third Generation” Sequencing—Nanopore Sequencing?
Tiny holes (nanopores) are made in specialized membranes through which an electric current is passing
This causes disturbances in the membrane’s electrical field
The different shaped nucleotides cause different specific types of disturbances, thereby allowing the machine to read the sequence of nucleotides in a DNA fragment as it gets pulled through the nanopore
What Does Real-Time PCR Allow For Quantitative Studies?
Real-time PCR determines the yield during the log phase of the PCR, and compare the sample’s yield to the yield of samples with known quantity of template
You can combine this with reverse transcription-PCR to do quantitative gene expression studies

What is Positional Cloning?
Sometimes called “forward” genetics–begins with a phenotype and finds the region where the responsible gene(s) lie(s)
You begin by identifying a region of a chromosome you will search in
Linkage analysis identifies a chromosome region in which a relevant gene resides
Association studies identify the gene alleles that are associated with the disease/trait of interest
Begin with either the marker(s) that define the linkage region, or the SNP, and identify a clone that has that sequence in it
Sequence the ends of the clone, and screen other clones to find some that overlap the original clone
What is Chromosome Walking?
Chromosome walking involves identifying and sequencing overlapping clones

How do you induce mutations to discover genes involved in diseases/traits?
“Reverse” genetics begins first by mutating the gene, then studies the phenotypic consequences of these mutations
Some studies induce mutations randomly throughout the genome with X rays or chemical mutagens and determine the phenotypic consequences
Others induce specific mutations in specific sites—for example by using the site you want mutated as a PCR primer binding site and making a primer that has the desired mutation it in
Describe How Genomewide Mutagenesis Allows Researchers To Identify Gene Functions
You can treat an organism with a mutagen (ex. ethylmethylsulfonate, EMS) that makes random mutations throughout the entire genome
You cross the organism with a pure-breeding wild-type organism; the F1 generation will show you the dominant mutations
How do you detect Recessive Mutations in Genomewide Mutagenesis
To detect recessive mutations, you cross F1 offspring that have wild-type phenotype with untreated pure-breeding wild-type animals to produce the F2 generation, some of which will be carriers of recessive mutations in genes of interest
You then backcross F2 offspring with their F1 parent
Any offspring that show a variant phenotype are homozygous for recessive mutations in a gene of interest






