Midterm 1 Flashcards
(41 cards)
define haploinsufficiency
- when one functional copy of the gene is NOT enough to provide enough proteins for normal function
define fertilization
- fusion of sperm and egg to form a zygote
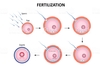
define cleavage
- Rapid cell divisions dividing the zygote cytoplasm into many progressively smaller cells called Blastomeres
- NB: no increase in volume
- by the end of cleavage, the blastomeres would have formed a sphere called blastula
what are the main patterns of cleavage?
- Holoblastic: complete cleavage; the entire egg is divided into smaller cells (mammals)
- Meroblastic: only part of the egg is divided and destined to form the embryo and the other part becomes the yolk (source of nutrition) (chicks)
- the yolk rich pole is called the vegetal pole and the embryonic side is called the animal pole
- Centrolecithal: is a form of meroblastic cleavage in which the yolk is in the center of the egg (fruit flies)

define gastrulation
- period of extensive cell rearrangement
- the stage in which the three germ layers form that will interact to form the organs

what are the requirements to meet to make gastrulation happen?
- EMT: epithelial to mesenchymal transition
- different types of cell movement
- formation of germ layers
- formation of the notochord
describe EMT

describe the different types of cell movement
- Invagination
- Involution
- Ingression
- Delamination
- Epibody

How are cells divided during gastrulation?
- ectoderm
- mesoderm
- endoderm
- germ cells

why is the formation of the notochord important?
- The notochord induces the formation of the nervous system
- the notochord forms from the mesoderm

define organogenesis
- resulting cell differentiation from chemical signals exchanged between cells of the germ layers
- ends in the formation of different organs
define the larval stage
A sexually immature organism
in order to become sexually mature, the larva needs to undergo metamorphosis
the adult stage is the shortest stage with the sole purpose of reproduction( life may end after giving birth)
define specification
- when a cell is committed to a given fate but this commitment can be reversed by changing the environment
- refers to a cell’s ability to differentiate in a neutral environment
- to check isolate cell and culture in a basic medium–> cell should differentiate

define cell determination
- The commitment to a given fate is irreversible
- the cell will differentiate to it’s committed fate regardless of its location ( a different part of the embryo or in culture)

define differentiation
- development into a specialized cell type
- At this stage, the cell is overtly a certain type at a biochemical and a functional level
describe the 2 types of specification
- Autonomous speciation: blastomeres acquire determination factors (determinants) from the egg cytoplasm as they divide. Nearby cells usually differentiate into the same thing since they are located next to each other. Even when isolated, the cells divide and become ciliated in time
- Conditional specification: specification comes from signals released from nearby cells. An intact 4-cell embryo from a pluteus larva can be isolated into 4 and produce a full but smaller organism.

describe syncytial specification
syncytium: a cytoplasm containing many nuclei. Cell membranes haven’t formed between nuclei.
* the cells already contain determinants but in this case, determinants can diffuse and influence nearby nuclei

define morphogen
- A long-range signaling molecule that forms a concentration gradient in the embryo and the specification/differentiation of a cell depends on the concentration of the molecule

describe histone tail modifications
- methylation: condense nucleosomes more tightly together blocking access to promoters
- acetylation: loosens nucleosome packing exposing DNA to RNA polymerase II and transcription factors
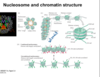
describe methylation of H3 tails
- methylated lysines on the H3 histone tail can be bound by either Polcyomb- a transcription repressing protein
- OR by trithorax proteins that stimulate transcription
- the binding or polcyomb or trithorax depend on the lysine position on the H3 tails
- methylation of L4,38 and 79 are associated with gene activation
- methylation of L9 and 27 cause repression

describe the central dogma of biology
- transcription of DNA results in a nuclear RNA sequence containing a 5’cap, 3’ polyA tail and untranslated regions
- Nuclear RNA is processed into messenger RNA through the removal of the introns
- The mRNA is translated by ribosomes
- The protein is usually inactive until post-translational modifications are done on it

what are the 3 types of transcription factors? and their domains
- TFs that bind to promoters
- TFs that bind to enhancers
- TFs that link other TFs( promoter TF to enhancer TF)
- DNA binding domain
- trans-activating domain: binds other proteins that act on inactive genes
- protein-protein interaction domain: allows the formation of a complex (linking TF or TF-promoter)
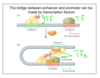
define an enhancer
- noncoding DNA sequence that is bound by TFs and activates transcription of a specific gene cis (downstream)
- an enhancer can be up to one million bps upstream from the promoter
- separate enhancers control the expression of a developmental gene in different tissues
- mutation in an enhancer causes tissue-specific phenotypes
how to identify the tissue-specific enhancer?
- fuse suspected enhancer with a reporter gene and check for expression
- ex of reporter genes: B-galactosidase and GFP protein