Structure of nucleic acids and genome organization Flashcards
(34 cards)
replication of DNA is catalyzed by which enzyme.
requires
DNA strand grows in which direction?
DNA polymerases
- requires
- template
- ssDNA
- substrates
- dATP, dGTP, dCTP and dTTP
- template
- growth in the 5’->3’ direction
differentiate from prokaryote and eukaryote origin of replication
- similar
- _melting -seperating of the DNA_
- BIDIRECTIONAL synthesis
- differences
- euk
- multiple OriC’s
- RNA primers are removed by RNAse H,
- euk
what enzymes are involved in folowing, for prokaryotes
- melting the DNA
- keeping the two strands apart?
- unwinding the double helix
- DnaA
- protein binds and melts dsDNA
- DNA helicase
- unwinds double helix
- ssDNA-binding
- proteins bind to ssDNA
- keep strands separated
- they protect ssDNA from nucleases
What is the generation/solution to a supercoil?
- generation
- unwinding of the DNA forms positive supercoilds ahead of replication fork.
- These can interfere with helicase function
- solution
- topoisomerase removes the supercoil, by nicking one side, rotating the DNA and ligating
DNA
- DNA polymerase read DNA in the ______ direction
- DNA polymerase synthesizes DNA in the ______ direction
- what is the consequence of this action
- describe the two
- direction and type of synthesis
- DNA polymerase
- reads in the 3’->5’ direction
- synthesizes in the 5’->3’ direction
- two types of strands form b/c the DNApol can only perform the above directions
-
leading strand
- being copied i nthe direction of replication fork
- continuously synthesized
-
lagging strand
- being copied in the direction away from the replication fork
- okazaki fragments:small fragments of DNA
- discontinuously synthesized
-
leading strand
describe enzymes involved in initiation and elongation of DNA
- DnaA- melts DNA
- SSB-keep from reanealing
- helicase - unzips DNA
- leding strand
- continuous polymerization by DNApol3
- lagging strand
- primase establishes RNA primers
- DNApol3 - starts at the RNA primer and continues until it reaches the next RNA fragment
- DNA pol1 - locates the space between DNA and RNA, replaces the RNA primer with DNA
- Ligase forms P bond between 3’ and 5’ end
what are the functions of DNA ligase?
- Form bond in the okazaki fragments, between 3’ and 5’ phosphates.
- DNA repair and recombination
- recombinant DNA etchniques
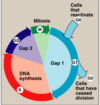
explain each phase of cell replication
- G0
- non replicative
- G1
- period preceding replication
- S
- replication occurs during
- mitosis
- seperationg of the cells
- G2
- chromosomes are formed
When the RNA primer is removed from extreme 5’ end of the chromosome, what the consequence of this? solution?
shortening of tthe new strand can result in loss of genetic information and is associated with aging
- soluution
- telomeres
- region of highly repetitive DNA at tthe end of chromosomes
- 300-600 base pairs in yeast
- function
- buffer consumed during cell division
- protect the ends from nucleases
- limit** cells to a fixed **number of divisions, in most somatic cells
- replaced by telomerase
- this enzyme is active in cancer cells, germ and stem cells
- telomeres
describe the mechanism of telomerase
- telomerase - ribonucleoprotein complex composed of proteins and RNA
- RNA-dependent DNA pol
- contains a telomerase RNA component
- which is complementary to a repetitive sequence in telomere
- mechanism
- uses hte 3’ end of DNA as a primer and TERC asa template to extend 3’ end of DNA
- once a single-stranded portion of telomere end is extended an RNA primer is produced to fill a single-stranded portion of chromosome

describe the affect of shorter telomeres
shorter telomeres affect hematopoiesis.
decreases the stem cell supplies

Describe 3 disease associated with TERC
- telomerase RNA component
- Aplastic anemia
- autosomal dominant dyskeratosis congenita
- familial MDS-AML
Tm depends on
Tm = melting point of DNA, temperature at which 50% of the dsDNA is separated
GC content
- higher GC= higher Tm
describe nucleases
- function
- types
nucleases
- phosphodiester bonds are recognized and cleaved
- types
- specific
- can recognize and bind a specfic sequence of nucleotides
- nonspecific
- can cleave nucleic acids at random positions
- specific
describe the following test
- FISH
- sounthern blot
- northern blot
- PCR
- FISH
- fluorescent in situ hybridization, used to detet specific sequences in genome
- southern blot
- DNA detection specific sequences after separation in the gel
- Northern blot
- RNA detection specific sequences after separation in the gel
- PCR
- is used to ampligy specific DNA sequences
describe the organization of geneomes in the following
- viruses
- prokaryotes
- Viruses
- lack complete systems for replication, transcripttion, and translation. hijack hosts system for their reprodcution
- prokaryotes
- circular c’some
- DNA-protein complex is located in nucleoid
- these organism lack a defined nucleus
-
plasmid
- small circular DNA molecules
- replicate autonomously, outside host genome
- used for genetic engineering and lateral gene transfer
describe the DNA associated with histones to form chromatin- 2 types
- Diffuse euchromatin
- transcriptionally active
- condense-hetterochromatin
- transcriptionally silent
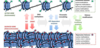
before mitosis DNA is replicated and forms two _____ ______.
What is the name of the region that microtubules attach to?
What about mtDNA?
DNA is replicated and form two sitster chromatids
centromer is the region of chromosome to which the microtubules of the spindle attach during cell division
mtDNA is independent of host cell division, must tkeep up and move on its own, nherited maternally
explain the CFTR gene: 7q31.2
- 7=chromosome number
- q=long arm
- 31= region, band
- 2=sub band
what is the basic unit of heredity in all living organisms?
what is the name of a specific spot on that unit?
alternative version of a this unit are called?
gene = the basic unit of heredity in all living organisms
genetic locus is a position of a gene on a c’some
alleles= alternative versions of agene.
homologous chromosome-each gene has a matching gene on a complimentary c’some (one from mom and one from dad)
how many c’somes do the following have ?
- haploid cell
- diploid cell
- haploid
- 23 c’somes
- diploid
- 46 c’somes
- 22autsomal pairs
- each composed of two homolous c’somes
- 2 sex chromosomes
- 22autsomal pairs
- 46 c’somes
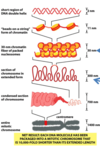
explain the structure used to compact DNA.
what are the superstructures?
histones
- RNA protein core allowing supercoiing of DNA
- DNA + core = nucleoid
- basic proteins (+ charge)
- five major classes
- H2A,H2B,H3,H4, and H1
what makes up the nucleosome core?
describe the structures and super structures.
- wrapping -140 bp of DNA around nucleosome clore
- nucleosome core= 2 of tthe following (H2A,H2B,H3, and H4)
many nucleosome cores coiled up generate a solenoid (aka 30nm fiber)
function= controlling transcription

list the composition and organization levels involved with DNA
- DNA-double helix
- chromatin
- 30nm=solenoid
- chromosome