Transcription and RNA processing I Flashcards
What are the transcription differences between eukaryote and prokaryote cells?
In prokaryotes, ribosomes have immediate access to mRNA and translation begins on the nascent mRNA chain while transcription is still in progress. In eukaryotes, mRNA synthesized in the nucleus must first be transported to the cytoplasm before it can be used as a template for protein synthesis. DNA in eukaryotes is packaged with proteins that can impact the ability of RNA polymerases to transcribe. DNA packaged in this manner is called chromatin. Chromatin that is wound tightly together makes it difficult for RNA polymerases to access the DNA.

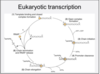
Eukaryotic transcription diagram:

Describe RNA polymerases.
All RNA polymerases have 9 conserved subunits, 5 of which related to bacterial RNA polymerase subunits.
Transcription factors required are distinct from the RNA polymerase.
RNA polymerases do not require a primer, read the template strand in the 3’-5’ direction.

What are the RNA pol2 promoters?
Promoters contain several sequence elements surrounding transcription sites
TATA box located 25-30 nucleotides upstream of transcription start site (resembles the -10 Pribnow element of bacterial promoters) – only present in 10-20% of RNA polymerase II promoters

What are the first two steps of forming pre initiation complex of transcription by RNA pol2?
TBP – TATA-binding protein
TAFs – TBP-associated factors
CTD – C-terminal domain
TF – transcription factor

What are 3 and 4th steps of forming preinitiation complex of rna transcription?
TBP – TATA-binding protein
TAFs – TBP-associated factors
CTD – C-terminal domain
TF – transcription factor
TFIIB acts as a bridge to join RNApol2-TFIIF to the complex.
TFIIH helicase subunits are XPB and XPD proteins – also required for nucleotide excision repair.
The preinitiation complex is composed of RNApol2 + five general transcription factors that are minimally required for initiation of transcription in vitro

What is the final steps of initiation of transcription?
Mediator complex stimulates basal transcription, and also plays a role in linking the general transcription factors to the gene-specific transcription factors that regulate gene expression
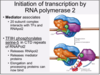
What are the two models of transcrition initiation by RNA pol2?
Two distinct mechanisms of formation of preinitiation complex:
(A) the stepwise binding of GTFs, pol II, and Mediator (Med)
(B) binding of a single multiprotein complex composed of pol II, Med, and the GTFs (Haloenzyme)
ORF – open reading frame

What enzyme transcribes rRNA genes and explain? (except the 5s)
The same gene encodes 5.8S, 18S, 28S rRNAs. 5S rRNA is encoded by a separate gene.
UBF – upstream binding factor
SL1 – selectivity factor 1
I don’t expect you to know the TFs here

What type RNA’s does RNA polymerase III transcribe?
snRNAs – small nuclear RNAs involved in splicing and protein transport
I don’t expect you to remember specifics about these promoters other than their unusual locations

Where is rRNA transcribed and processed?
Cells require large numbers of ribosomes to meet their needs for protein synthesis. Actively growing mammalian cells contain 5-10 million ribosomes that must be synthesized each time the cell divides.
5S rRNA is transcribed outside of the nucleolus.
The nucleolus organizes around the chromosomal regions that contain the genes for 5.8S, 18S, and 28S rRNAs, which are called nucleolar organizing regions.
How is rRNA processed, what are the diferent species of rRNA for eukaryotes and prokaryotes?
Eukaryotes have four species of rRNAs: 5S, 5.8S, 18S, 28S.
The 5.8S, 18S, and 28S are derived by cleavage of a single transcript.
18S > 40S ribosome
5.8S, 28S (and 5S) > 60S ribosome
Bacteria have three rRNAs: 5S, 16S, 23S. These are equivalent to the 5S, 18S, and 28S eukaryotic rRNAs and are also derived by cleavage of a single transcript.

What chromosomes are the genes that code for the rRNA species found on and describe this?
To meet the cell’s demand for transcription of large numbers of rRNA molecules, cells contain multiple copies of the rRNA genes. The human genome contains about 200 copies of the same gene that encodes 5.8S, 18S, 28S rRNAs organized in tandem arrays on 5 different human chromosomes (13, 14, 15, 21, 22). It also contains approximately 2000 copies of the gene that encodes 5S rRNA, organized in a single tandem array on chromosome 1
To meet the cell’s demand for transcription of large numbers of rRNA molecules, cells contain multiple copies of the rRNA genes. The human genome contains about 200 copies of the same gene that encodes 5.8S, 18S, 28S rRNAs organized in tandem arrays on 5 different human chromosomes (13, 14, 15, 21, 22). It also contains approximately 2000 copies of the gene that encodes 5S rRNA, organized in a single tandem array on chromosome 1

What are snoRNAs and what do they do?
Small nucleolar RNAs (snoRNAs) complex with several proteins to form small nucleolar ribonucleoparticles (snoRNPs). Nucleoli contain more than 300 proteins and 200 snoRNAs that function in pre-rRNA processing.
2 families of snoRNAs that associate with different proteins to catalyze methylation or formation of pseudouridines. snoRNAs contain ~15 nucleotides that are complementary to 18S or 28S rRNA – this region is the site of base modification.

Describe important aspects of tRNA processing.
Ribozyme – an enzyme in which RNA rather than protein is responsible for catalytic activity; RNA is active without the protein
Some tRNAs already have the 3’ CCA encoded.
Some tRNA and rRNA genes have introns that are removed by splicing

What percent of tRNA bases are modified and what functino do the modified bases play?
Functions of modified bases – most unknown, some play important roles in protein synthesis by altering base-pairing properties of tRNA molecule and stabilize hairpin loops



