2/3 DNA to Protein: Transcription and Translation Flashcards
(32 cards)
What are microRNA (miRNA) and what are the functions?
- small non-coding RNA molecule (~22 nucleotides) that function in RNA silancing and post-transcriptional regulation of gene expression.
- Gene silancing in plants and animals
What are the steps for microRNA biogenesis?
- pre-miRNA folds into particular structure w/ hairpin loop
- pre-miRNA gets trimmed (microprocessor formation)
- Nucleus → cytoplasm
- Dicer cuts off hairpin loop → miRNA duplex (miRNA + mRNA)
- Dicer (RISC coplex) further trims miRNA and pairs it with mRNA transcript
- *miRNA needs to be single stranded to be activated
Can miRNA’s target mutliple genes?
- many miRNAs can target one gene or….
- one miRNA can target many genes
What does miRBASE archieve?
- information about miRNAs including clinical significance
How are rRNAs modified in the nucleolus before they leave the nucleus?
- 2’ O methylated nucleotide
- pseudouridine (rotation of side chains)
- Ribosomal RNAs have more than 100 of each type of modification
- snoRNAs determine the sits of modification by base-pairing to complementary sequences on the precursor rRNA. The snoRNAs are bound to proteins, and the complexes are called snRNPs.
- SnoRNPs contain the guide sequences and the enzumtes that modify the rRNA.

What happens in the nucleolus?
- Assembly of the ribosomal complex
- Assembly of the telomerase complex
- its not membrane bound.
- pre and post processing - rRNA and the proteins needed to asssemble the ribosome.
How does the Nucleolus produce Ribosomes?
- Precursor rRNA is packaged with many proteins
- → Immature large & small ribosomal subunits
- Final processing doesn’t occur until the immature formsare shipped through the nuclear pore to the cytoplasm.
What happends at the location of cajal bodies?
- Cajal bodies are where most snoRNAs are assembled into snRNPs

What is the tRNA structure like?
Where is the acceptor stem & anticodon within the structure?

What is the wobble position?
- Some tRNAsonly requrie accurate base-pairingonly at the first two positions of the codon and can tolerate a mismatch (or wobble) at the third position.
- Wobble position makes it possible to fit the 20 amino acids to their 61 codons with as few as 31 kinds of tRNA molecules.

Why do tRNAs splice introns out by a specialized set of proteins and not the spliceosome?
- Trimming and splicing require the tRNA to be folded properly and will not proceed if the tRNA is misfolded.
- The splicesome is not set up to take into account 3D structure
- So specialized proteins evolved just for tRNAs
What happens before tRNAs exit the nucleus?
- tRNAs are covalently modified before they exit from the nucleus.
- ~10% of tRNAs are modified.
What is aminoactyl-tRNA synthetase?
- aminoactyl-tRNA synthetase helps ensure that the correct amino acid is coupled to the tRNA
- The tRNA is an adapter whose anticodon pairs with the codon in the mRNA to identify the correct amino acids to add to the growing peptide.
- 3D interaction between amino acids of protein and nucleotides of tRNA.
*

What is the fundamental reaction of protein synthesis?
- the formation of a peptide bond between the carboxyl group at the end of a growing peptide and the amino group on the incoming AA.
- Formation of the peptide bond is energetically favored.
Prokaryotic vs Eukaryotic ribosomes
- Bacterial Ribosomes
- 70S
- 50S, 30S
- 70S
- Eukaryotic Ribosomes
- 80S
- 60S, 40S
- 80S
What binding sites does a ribosome have?
- each ribosome has one binding site for mRNA and 3 sites for tRNA
- A, P and E.
- A= amino actl tRNA
- P= peptidyl-tRNA site
- E= exit
What are the four steps in protein synthesis?
- New tRNA binds to A site pairing with codon
- Carboxyl end of growing peptide is released from tRNA in P site. Peptide bond formation between the previous AA added and the new one. tRNAs are in P site and A site
- Large subunit moves along mRNA held by small subunit, shifting tRNAs to P and E sites on small subunit
- Small subunit moves 3 nucleotides along the mRNA and ejects the tRNA in the E site.
What do Elongation facotrs do in protein synthesis?
- elongation factors drive translation forward and improve its accuracy
What is the primary role of proteins?
- to stabilize the RNA core.
Is Ribosome a Ribozyme?
- Yes,
- RNA is the enzyme that catalyzes the formation of the peptide bones and . . .
- 2/3 of a ribosome is RNA.
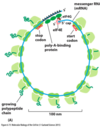
How do nucleotide sequences in mRNA signel where to start protein synthesis?
- Initiation of protein synthesis requires the presence of several translation initiation factors. The poly A tail must be bound by poly A binding proteins that interacts with eIF4G to ensure that both ends of the mRNA are intact
- 2 GTP hydrolysis steps provide the energy needed.
- A consensus sequence flanking the start codon tells the ribosome to begin translation on this AUG. If the flanking nucleotides differ much , the ribosome will move to the next AUG.This results in alternative starting proints for the protein.
How is translation stopped?
- binding of a release factor to an A site containing a stop codon causes termination of translation.
- The peptide is released and in a series of reactions the ribosomal subunits dissociate and are then ready to initate translation again.
Why are proteins made on Polyribosomes?
- Upon completion of translation, the ribosomal subunits are near the 5’ end of the mRNA because of the association of the 5’ and 3’ ends of the mRNA. Thus they can restart quickly.
How do inhibitors of prokaryotic protein synthesis useful in antibiotics?
- Fungi produce many antibacterial componds that exploit differences in ribosomal subunits.
- Fungi produce all kinds of small molecules that inhibitor bacterial protein synthesis