W12 Flashcards
(31 cards)
Types of isomerisation in complex ions

Coordination isomersation in complex ions
Same formula but formula of compex ion is different

Linkage isomers
Complex ion has same formula but ligand is ambidentate and can bind through different donor atoms
cyanide
M:CN-
M:NC-
cyanido-κC
cyanido-κN
thiocynate
M:SCN-
M:NCS-
thiocynato-κS
thiocynato-κN
nitrite
M:NO3-
M:ONO2-
nitrito-κN
nitrito-κO
Colours
Isomers have different colours - even linkage isomers
Geometric isomers
Atoms of groups of atoms assume different ligand binding positions in the complex
- Cis/trans isomerisation: two constitutent on the same/different side
- Fac/mer isomerisation: (octahedral complexes) three constituents making up a face/meridian
Cis/trans isomerisation

Fac/mer isomerisation
1 axial, 2 equatorial atom
2 axial, 1 equatorial atom
Takes up 1 octahedral face
Cuts through meridian

Optical isomerisation
What is it
naming
example
Nonsuperimposable mirror images called delta and lambda
For 2x bidentate ligands in octahedral structure:
- Delta: 1x (equatorial to axial) 1x (equatorial to equatorial)
- Lambda: 2x (equatorial to equatorial)
Triscatecholates metal complexes have optical isomerisaion
Sidephores
Chelates that can bind to iron irons very strongly
Are secreted by micro-organisms
Examples
- catecholates (Fe3+)
- enterobactin (Fe3+)
- hydrogen phosphate (Fe3+)
- haem group (Fe2+)
Catecholate
Bidentate, dianionic/divalent chelating ligand whne bound to metal
Each donor atom (oxygen) is uninegative
Formed by deprotonating catechol

Triscatecholatoferrate(III)
- 3 catecholates bind to one Fe(III)
- Overall 3- charge
- Very high equilibrium constant

Enterobactin reaction
- Compound produced by aerobic bacteria
- Forms [FeEb]3- = lipophilic - able to transfer Fe into cells
- Cyclic polyester forms amide links to 3 catechol-type ligands
- It becomes a hexadenate, hexavalent ligand, binding to Fe(III) in the middle
- Overall 3- charge

Enterobactin reaction in aerobic bacteria
Triscatecholato is LESS stable than enterobactin
Because of chelate effect:
- Enterobactin is a hexidentate ligand while triscatecholatoferrato(III) is three bidentate ligands
- If one of the tricatecholato bonds to metal is broken, it is lost from the complex (because of entropic driving force) but in case of enterobactin a dissociated catecholate will reattach itself (no entropic driving force)

Hydorgen phosphate
- Hydrogen phosphate is a bidentate ligand that binds to Fe3+
- Both O- act as donor atoms
- forms [FeIII(H2O)4(HPVO42-)]+
- iron complex stable at pH 2 of stomach
- used to supply blood to transferrin
Naming coordination complexes
Order
- Ligands are named in alphabetical order before metal ion
- Cation is named before anion
Ligands
- Anion endings change
- Ide -> ido
- Ate -> ato
- Di, tri, tetra, penta, hexa indicate number of “simple” ligands
- Bis, tris, tetrakis are used to indicate number of “complex” ligands
Metal
- Oxidation state of metal ion is indicated by roman numerals
- For metals in anionic complexes: sometimes latin name is used
- Ferrate Fe
- Cuprate Cu
- Platinate Pt
- Plumbate Pb
- Argenate Ag
- Aurate Au
- Stannate Sn
Secondary structure of proteins
Alpha helix
Beta pleated sheet
Structure: Helical
- Intra-chain hydrogen bonding
- C=O…H-N
- All side chains point away from helix
Function: Gives elasticity to proteins
- Wool, hair: alpha keratin
- Tendons: collagen
Structure:
- Inter-chain hydrogen bonding
- C=O…H-N
- Side chains point alternatively above and below plane of sheet
Function: Gives rigidity to proteins
- Silk

Transferin structure
-
2 identical subunits of 40k Da
- Contains both beta and alpha sheets
- Held together by:
- Ionic bonds
- H-bonds
- Etc.
- Fe binding constant: 1026

Transferrin structure
-
Transport iron INTO cells (<1% of body’s total Fe content)
- Travels in blood
- Docks with transferrin receptors on cell surfacers
- Taken into cell, drop in pH causes release of Fe3+
- Demetalled protein is returned to outside of cell, process continues
-
Bactericidal
- Transferrin is in mucus, egg whites, milk
- Scavenges for iron - deprives bacteria of it
- Passes Fe to ferritin (iron storage protein)
- Can remove Fe3+ from phosphate and citrate
How does transferrin carry iron?
Draw
- Fe3+ is buried deep into transferrin
- Coordination environment contains anionic and a neutral ligands
- Monodentate: 2 tyrosines(-), 1 histadine, 1 aspartic acid(-)
- Bidentate: 1 carbonate

Ferritin structure and function
Structure
- Rust coated with soluble protein
-
Protein coat or shell:
- Subunits are roughly cylindrical, 163 amino acids
- 24 subunits form hollow sphere with large diameter (120 angstroms)
-
Rust core
- 75 angstroms across
- All iron is Fe3+
- Mainly FeO(OH) with phosphate ligands
-
Core-protein interface
- Fe ions enter through channels at corners
- Ion channels are surrounded by oxygens
- Helps polar iron complexes pass through
Function
- Store iron (takes it from transferrin)
O2 binding proteins
Uses Fe2+ in heme functional unit
- Haemoglobin (blood): transports oxygen
- Myoglobin (muscles): stores oxygen
Haem unit
- Based on a deprotonated porphyrin ligand with Fe2+ at centre
- Square planar, tetradentate, dianionic/divalent ligand
- Aromatic so flat
- Has two propanato groups attached to porphyrin ring
- Bulky
- Negatively charged
- Hydrophilic
- Also has methyl and vinyl subunits (not shown)
- Oxygen binds to open face opposite the two propanato side groups
- When deoxygenated, water binds to open face opposite the propanato side groups
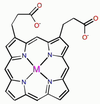
Structure of a protonated porphyrin ligand
- Aromatic (26 pi electrons) with many resonance forms
- Flat, square planar coordination geometry

Myoglobin structure, structure of haem group binding site, function
Structure
- Monomeric (only one protein subunit)
- 8 alpha helices fold around 1 heme unit
- Helices are held together by:
- Hydrogen bonding
- Covalent (disulfide bridges)
Structure of Haem group
- Haeme group is held to protein side chain through histidine side chain
- through nitrogen on imidazole side group binding to propanato)
- Oxygen binds to site opposite (trans) to histidine group
-
Crowding of coordination site allows only very small molecules to have access to Fe(II)
- Oxygen is small enough to pass through and bind
Function
- Stores oxygen in tissues (muscles)
- Myoglobin converted to oxymyoglobin even at low O2