MCM_Final_TBL11 Flashcards
(39 cards)
Post-Translational Modification: Phosphorylation
- adds a phosphate, serine, threonine or tyrosine

Post-Translational Modification: Glycosylation
- attaches SUGAR
- usually at an “N” or “O” in an amino side chain

Post-Translational Modification: Ubiquitination
- adds ubiquitin to lysine residue of a target protein for DEGRADATION

Post-Translational Modification: SUMOylation
- adds a small protein SUMO (small ubiquitin-like modifier) to a target protein

Post-Translational Modification: Disulfide Bond
- covalently links the “S” atoms of two different CYSTEINE residues

Post-Translational Modification: Acetylation
- adds an acetyl group to an N-terminus of a protein or Lysine group

Post-Translational Modification: Lipidation
- attaches a lipid such as an (fatty acid) to a PROTEIN CHAIN

Post-Translational Modification: Methylation
- adds a methyl group at Lysine or Argenine residues

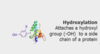
Post-Translational Modification: Hydroxylation
- attaches a hydroxyl OH group to a side chain of protein
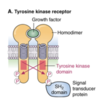
Process of Protein Excretion
-
membrane bound/excreted proteins are made** in the ribosomes located on the **membranes of the RER
- these proteins have a 1-36 N-Terminal signal peptide domain
- the signal peptide is recognized by a protein comlex called the SIGNAL RECOGNITION PARTICAL (SRP)
-
once the peptide moves past the ER membrane, the signal peptide is removed
- signal peptidase removes the signal peptide
- Proteins containing a signal peptide are called pre-proteins
Glycosylation
-
glycans add sugar to molecules
- can add monosaccharides (i.e transcription factors) OR complex branched polysacharides (i.e cell receptors)
- use activated donors like UDP-sugar OR GDP-sugar
-
attached at:
-
asparagine-linked (N-linked) oligosaccharides OR
- N-glycosylation occrs in RER
-
serine/threonine (O-linked) oligosaccharides
- O-glycosylation occrs in GOlgi
-
asparagine-linked (N-linked) oligosaccharides OR
- ex) Viral Spike Protein (Covid) binds ACE2 to gain entry into human hosts

Methylation
- can be added to either nitrogen** or **oxygen AA side chains
- mediated by methyltransferases & requires S-adenosyl methionine (SAM)
- uses:
- methylation = ↑ hydroPHOBicity
- TRImethylation = permanant charge shift
-
epigenetic regulation
- (ex) histone methylation: makes DNA unaccesible for transcription
SAM & Nutrition Connection
- SAM = methionine + ATP
- With the ACTIVATION of Vit. B12 (cobalamin), ATP donates adenosine
- after the transfer of methyl, SAM becomes S-adonosyhomocysteine (SAH)
- then hydrolyzed to form homocysteine and adenosine

N-Acetylation
- transfer of acetyl to lysine (REDUCES CHARGE)
- N-Acetylation is REVERSIBLE
- (methylation tends to be irreversible)
N-Terminal Acetylation
- initial Met is cleaved and
- N-acetyltransferases (NAT) enzymes use acetyl-coA to neutralize the N-terminal AA

Lysine Acetylation
- occurs on the side chain instead of the N-terminal
- ex) histone lysine acetylation ↓ charge = ↑ access of promotors, which allows transcription factors (TF) to COUNTERACT histone METHYLATION
- 2 enzymes:
- histone acetyltransferases (HAT)
- histone deacetylases (HDAC)
- 2 enzymes:
MyeloDysplastic Syndromes (MDS) and Acute Myeloid Leukaemia (AML)
- MDS & AML = man, immature BC = blood cancer
- caused by hypermethylation of β-catenin promoter
- Two Treatments:
- Azacytinide: stops doxy-Cysteine methylation
- Vorinostate: histone deacetylase (HDAC) inhibitor
Hydroxylation
- addition of OH to proline and others

Hydroxylation Disorder
- collagen = 25% of all protein in body
- collogen hydroxylation is done by prolyl-hydroxylase using Vit. C as a cofactor
- this rxn occurs at every 3rd AA in collagen to stabilize it’s triple helical structure
- therefore, LACK of Vit C = Scurvy (defective collagen formaton)…
-
symptoms of Scurvy:
- subcutaneous hemorrhage (bruising)
- aching bones/joints/muscles
- rigid position and pain
- recommended Vit C intake is 75-90 mg
Protein Lipidation
- attachment of lipid to proteins
- used to anchor proteins to membranes:
- organelle membranes (ER, Golgi, Mito)
- vesicle membranes (endosomes, lysosomes)
- plasma membranes
Major Types of Lipidation (4)
- C-terminal Glycosyl PhosphatidylInositol (GPI) anchors:
- N-terminal myristoylation, S-myristoylation (C14, Gly)
- S-palmitoylation (C16, Cys)
- S-prenylation (C15/C20, Cys)
GPI Anchors
- anchor cell surface proteins to plasma membrane
- due to the ER signal sequence, glycolipid is added to newly made protein
- they will be reversibly localized to cholesterol and sphingolipid rafts in plasma membrane for signaling platforms
- ex) when GCPR signaling anchor released from Phosphlipase C
N-myristoylation
- 14 C fatty acid on terminus
-
selective, reversible localization signal that binds only some proteins to membranes
- ex) Src-family kinases (i.e tyrosine kinases signals that regulate cell proliferation, differentiation, apoptosis, migration, and metabolism)
- enzyme: N-myristoyltransferases
S-palmitoylation
- description: C16 added to thiol of cysetine
- enzyme: palmitoyl acyltransferases
- function:
- permanant anchor of membrane proteins
-
on/off switch: to regulate membrane localization
- thioesterases can cleave Cys and the anchor
- strengthens other types of lipidation (i.e myristoylation OR farnesylation)